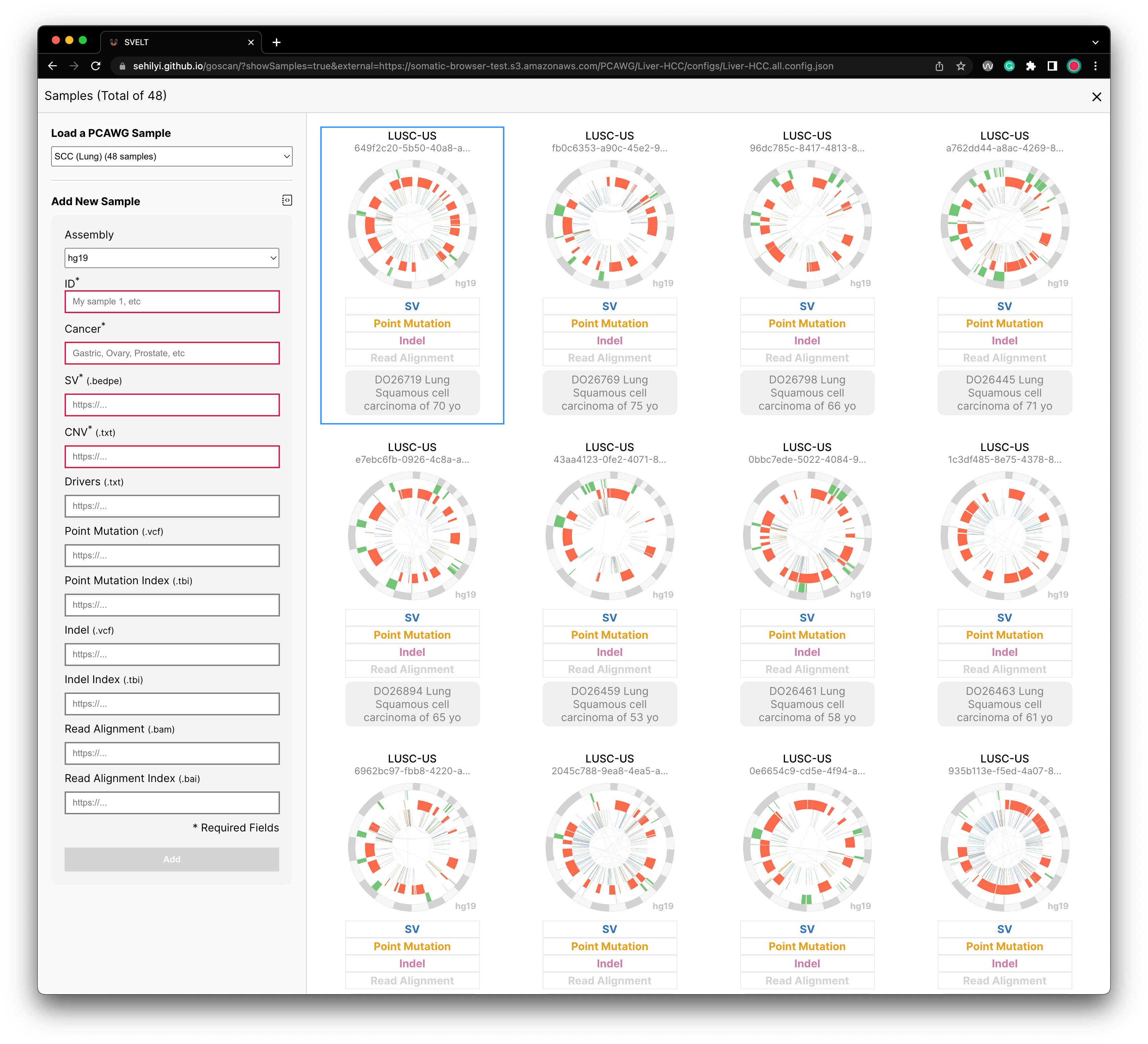
Loading Data Through Interface
You can load a small number of samples directly on the browser through the interface. In the cohort-level view, there is a side panel that enables you to add a new sample by providing metadata (e.g., cancer type) and file URLs (e.g., bedpe, txt, vcf).
 |
|---|
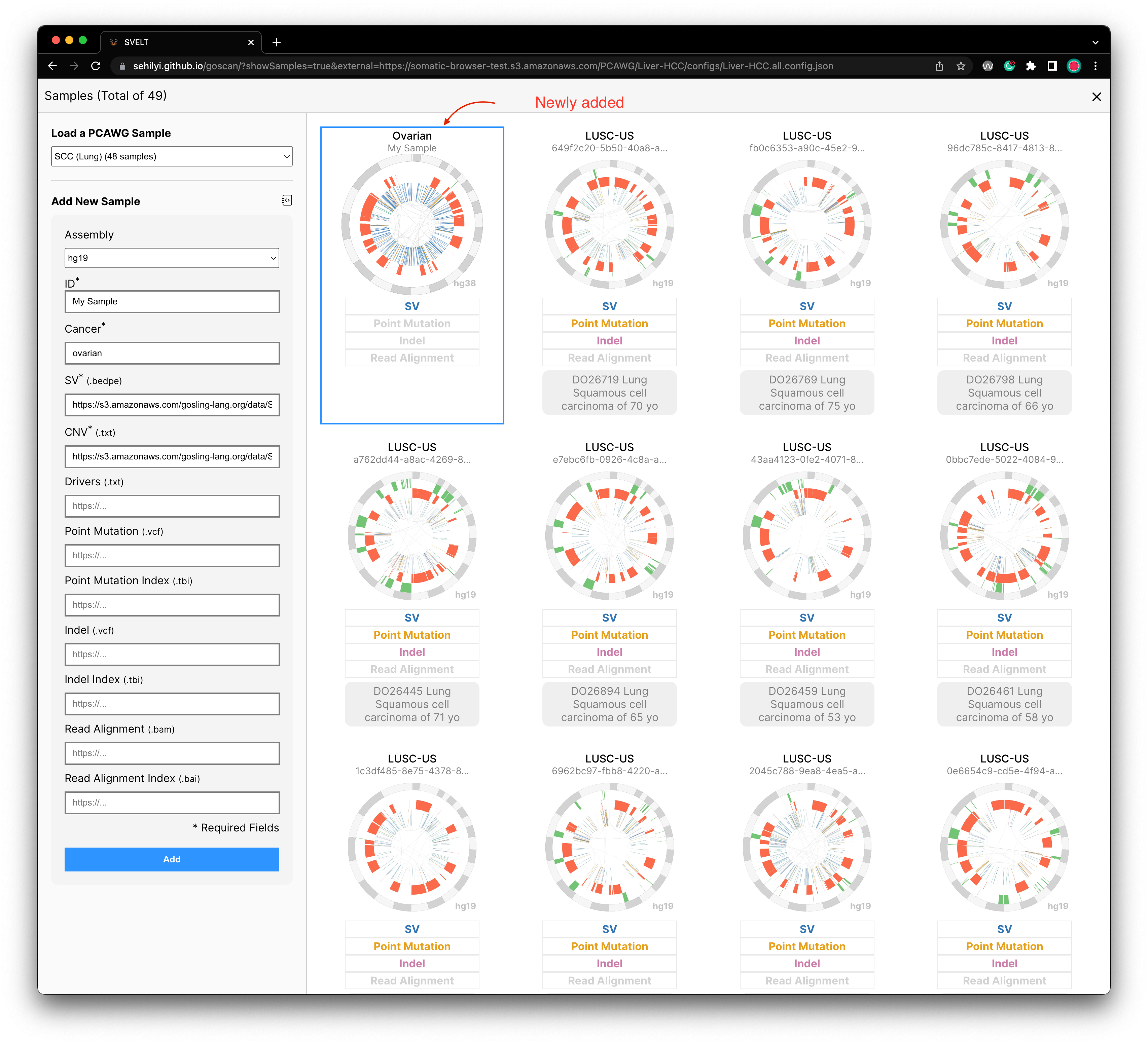
After providing all required information, you can click on the Add button on the bottom of this form. Once you click on it, you will be able to see a sample added as the first sample.
 |
|---|
help
To test the browser for adding a sample, you can use the following information:
| Fields | Contents |
|---|---|
| ID | 7a921087-8e62-4a93-a757-fd8cdbe1eb8f |
| Cancer | Ovarian |
| assembly | "hg19" |
| SV | https://s3.amazonaws.com/gosling-lang.org/data/SV/7a921087-8e62-4a93-a757-fd8cdbe1eb8f.pcawg_consensus_1.6.161022.somatic.sv.bedpe |
| CNV | https://s3.amazonaws.com/gosling-lang.org/data/SV/7a921087-8e62-4a93-a757-fd8cdbe1eb8f.consensus.20170119.somatic.cna.annotated.txt |
Details about each of the fields, as well as accepted files, are described in the following article.